Whole Genome Sequencing of
SARS-CoV-2: Progress Report
13/May/2020
Unless indicated otherwise, all content in this document is licensed for re-use under a
Creative Commons 4.0 International Licence © 2020 Institute of Environmental Science and
Research Limited
link to page 4 link to page 5 link to page 7 link to page 7 link to page 7 link to page 7 link to page 9 link to page 12 link to page 12 link to page 13 link to page 7 link to page 8 link to page 12 link to page 5 link to page 5 link to page 6 link to page 9 link to page 10 link to page 11

CONTENTS
Summary ............................................................................................................................. 2
International linkages ......................................................................................................... 3
Analysis of known outbreaks and clusters ....................................................................... 5
Genetic linkage of samples to known cases or outbreaks ................................................ 5
Cases locally detected, source unknown ..................................................................... 5
Outbreak analyses ....................................................................................................... 5
Genetic linkage to known outbreaks............................................................................. 7
Identification of transmission patterns ............................................................................ 10
Three different transmission chains identified within one outbreak ............................. 10
Glossary ............................................................................................................................ 11
LIST OF TABLES
Table 1. Overview of sequencing results for cases with unknown source.............................. 5
Table 2. Overview of sequencing results for known clusters ................................................. 6
Table 3. Different transmission chains identified within the OB-20-108803-AK cluster ........ 10
LIST OF FIGURES
Figure 1. Overview of available whole genome sequencing data of COVID-19 cases
worldwide .............................................................................................................................. 3
Figure 2. Analysis of viral genomes related to recent travel to Iran ........................................ 4
Figure 3. Overview of the OB-20-108795-IN ......................................................................... 7
Figure 4. Overview of the OB-20-108823-AK ........................................................................ 8
Figure 5. Overview of the OB-20-108805-IN ......................................................................... 9
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 1

SUMMARY
This report summarises the findings from analyses of whole genome sequencing of COVID-
19 samples referred to ESR for sequencing since 01 February 2020. Epidemiological data
used in the analyses were collected from cases notified from 30 January 2019 to the
surveillance week ending 01 May 2020. Information is based on data recorded in EpiSurv as
at 04 May, changes made to EpiSurv after this time will not be reflected in this report. The
results presented may be updated and should be regarded as provisional.
In this first COVID-19 WGS report, given the small number of cases sequenced to date a
high proportion of the cases sequenced will not have genomic links to another known case
or cluster. More genomic linkages will be demonstrated in future reports as the proportion of
cases sequenced goes up and this will provide more robust data to analyse in combination
with the epidemiological findings. In the meantime caution should be used when interpreting
source and transmission chains, especially for cases not yet genomically linked to another
case or cluster, or for cases where the genomic and epidemiological findings appear
inconsistent.
As of 01 May ESR had received 623 COVID-19 samples for whole genome sequencing,
sequencing has been performed on 125 samples, resulting in 97 complete genomes of
sufficient quality for analysis and interpretation.
Analyses to date have assessed the likely international sources of infection for cases
diagnosed in New Zealand, whether cases with an unknown source can be linked to other
cases or clusters within New Zealand, and whether sequencing results support the
epidemiological findings used to assign cases currently linked to existing outbreaks and
household clusters.
International linkages
In all cases to date, the results from genomic sequencing support the epidemiological
information on the likely international source of infection.
Analysis of cases of unknown source and
known outbreaks/clusters
Genomic linkage of samples to known cases, clusters and outbreaks
• four out of six cases of unknown source were genomically linked to other known cases in
New Zealand
• 31 cases were confirmed to cluster with other cases in the outbreak they had been
assigned to based on epidemiological information
• 15 cases were genomically linked to known cases or clusters to which they had not been
linked using epidemiological data. These cases have been referred to PHUs for further
investigation.
Identification of differing transmission patterns within an epidemiological cluster
• Sequencing identified three different potential transmission chains within an
epidemiological cluster indicating that these cases were not all part of the same
outbreak.
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 2
link to page 5



INTERNATIONAL LINKAGES
Worldwide 10 different clades have been identifie
d and Figure 1 shows an overlay of the 97
complete genomes sequenced from New Zealand cases on the international sequencing
results. This shows the widespread international genomic links of New Zealand cases.
Figure 1. Overview of available whole genome sequencing data of COVID-19 cases
worldwide
The phylogenetic tree was generated using the Nextstrain analysis pipeline. Branches and
nodes are coloured according to their genomic clade. All New Zealand sequences are
highlighted with circles. The wide spread of New Zealand samples across the tree reflects
the high number of introductions into New Zealand through international travel.
In the early phases of the pandemic, the concordance between the countries of origin
identified by epidemiological data with the genomic data was investigated. The results
provided confirmation that the genomics workflow was working and could return robust
results within 48 hours. The analysed travel associated genomes were consistent with the
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 3

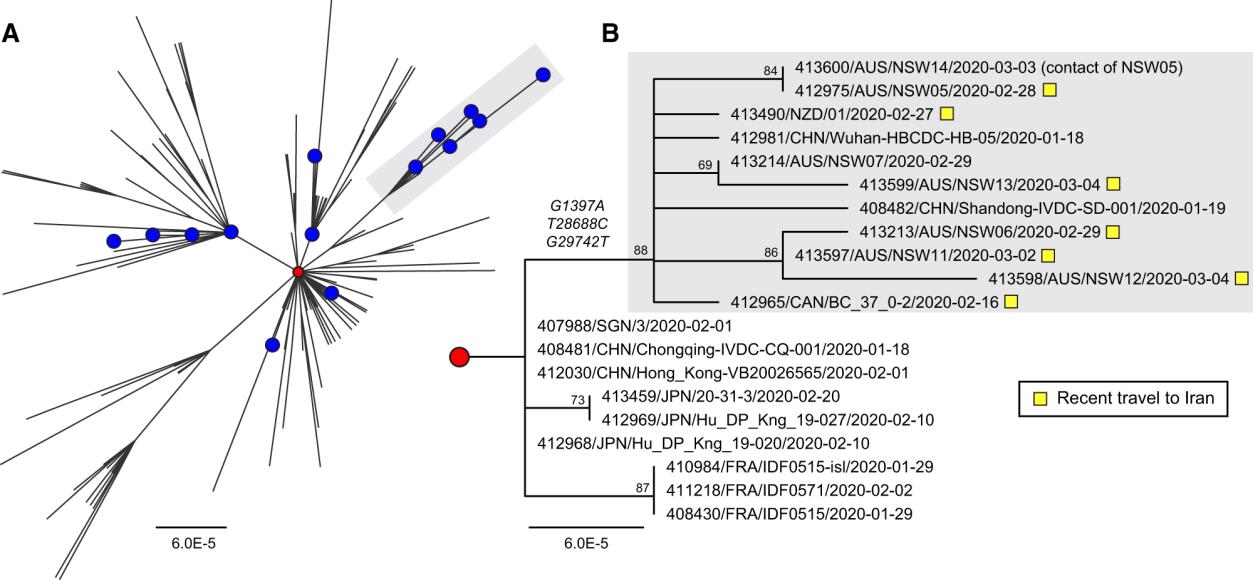
known travel history. In Figure 2 we provide an example of how these analyses were
performed.
Figure 2. Analysis of viral genomes related to recent travel to Iran
“Phylogenetic analysis of SARS-CoV-2 genome sequences highlighting a clade of imported cases from Iran. (A)
Global diversity of circulating SARS-CoV-2 strains including Australian sequences (blue circles, n = 19). The
prototype strain Wuhan-Hu-1 is shown as a red circle. An emergent clade containing cases imported from Iran is
highlighted with grey shading. (B) Sub-tree showing the informative branch containing imported Iranian cases
(highlighted with yellow squares) and defined by substitutions at positions G1397A, T28688C, and G29742T. Node
support is provided as bootstrap values of 100 replicates. For both (A) and (B), the scales are proportional to the
number of substitutions per site.”
Figure and legend reused under BY-NC/4.0 licence from:
Virus Evol, Volume 6, Issue 1, January 2020, veaa027,
https://doi.org/10.1093/ve/veaa027
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 4
link to page 8

ANALYSIS OF KNOWN OUTBREAKS AND
CLUSTERS
The findings in this section show examples of the value of sequencing to enhance and inform
epidemiological investigations and to understand chains of transmission.
GENETIC LINKAGE OF SAMPLES TO KNOWN CASES OR OUTBREAKS
Cases locally detected, source unknown
One of the useful impacts of genomics technology in communicable disease control is the
identification of genomic links between cases where no epidemiological link has been found
to a “source”. Understanding how the infection is spreading in the community, the chain of
transmission, is important when evaluating or planning containment measures. Therefore,
cases classified as ‘source unknown’ have been prioritised for sequencing to assist public
health responses.
ESR has sequenced six cases classified as “local y acquired, unknown source”. For four of
these cases genomics indicated potential links with other known cases or with an outbreak.
Below we present a summary of these findings and the subsequent epidemiological follow-
up.
Table 1. Overview of sequencing results for cases with unknown source
EpiSurv or lab ID of
Genomics region of
ESR ID
genomically linked case or
origin
outbreak
20VR****
USA
20VR****
20VR****
UK
Not yet detected
20VR****
Europe
Not yet detected
20VR****
USA, Australia, Europe
20VR****
20VR****
USA, Australia, Europe
20VR****
20VR****
NZ, Australia
OB-20-108795-IN cases
• 20VR****: There is a potential epidemiological link between these 2 cases as the linked
case was imported from USA and the source unknown case has work related contact
with many international travellers. However, as the onset date of illness was the same
day the imported case arrived in New Zealand, it is unlikely the genomically linked case
is the source for this case. It is more likely to have been an earlier arrival from the USA.
• 20VR**** & 20VR****: epidemiological data indicate that this was not a direct link but
more likely exposure to a common source through an imported or import - related case
who was travelling between regions while infectious. As more cases are sequenced
further links may be found.
• 20VR****: this person was resident in the region during the conference where the OB-20-
108795-IN originated. Extensive epidemiological follow-up has not found a direct link
between this case and a conference attendee, venue or travel.
Outbreak analyses
An overview of the outbreaks for which at least one case has yielded a high quality genome
is presented in
Table 2. Cluster consistency analysis can be performed when at least two
cases have been sequenced successfully. Inconsistencies noted are being followed up by
reviewing the epidemiology with the relevant public health unit (PHU) along with reviewing
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 5

the sequencing findings. Some apparent inconsistencies may be resolved once more cases
have had samples sequenced.
Table 2. Overview of sequencing results for known clusters
Samples epi-linked to
No OB* epi-
Comments on
Outbreak
link, cluster
observed
Outbreak ID in
No. at
No.
No. in
genomically
potential
EpiSurv
ESR
WGS**
OB
with OB cases
inconsistencies in
genomic No. samples
WGS
cluster
OB-20-108805-IN
80
8
8
3
See details below
20VR****
OB-20-108795-IN
28
15
14
2
Different clade. Other
possible epi-links
OB-20-108806-HN
9
2
2
0
OB-20-108787-WN 8
4
4
0
OB-20-108803-AK
6
4
2
1
See details below
Quite distant (2 mutations
apart). Epidemiological
information shows several
OB-20-108810-NA
6
2
2
0
generations from source
case and this case is a
later case in a subcluster
Additional case no known
OB-20-108799-WN 5
1
NA
1
epi-links with OB
Additional cases, one
imported, no epi-links
OB-20-108811-AK
5
1
NA
2
between these cases or
with OB
OB-20-108823-AK
4
1
NA
4
See details below
Additional case currently
epi-linked to another
OB-20-108820-AK
3
1
NA
1
cluster. PHU to review
other possible epi-links
OB-20-108814-AK
2
1
NA
0
OB-20-108813-CH
1
1
NA
0
Additional case imported
from USA, no direct epi-
links with OB cases but
OB-20-108837-TI
1
1
NA
1
OB cases exposed to
international travellers.
PHUs following up.
*OB – outbreak
**WGS – whole genome sequencing
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 6


Genetic linkage to known outbreaks
OB-20-108795-IN (Hereford conference)
Figure 3. Overview of the OB-20-108795-IN
Samples with epidemiological links are highlighted in purple.
Two specimens from one case have been sequenced for quality control purposes
Additional cases investigations:
Two additional cases cluster genomically with outbreak cases but neither were epi-linked to
the outbreak:
• 20VR**** was resident in the area during the event but additional epidemiological follow
up has not been able to find a direct link with other cases epi-linked to the outbreak or to
the conference venues, for either this case or their flatmate, who was also a lab-
confirmed case with onset within 24 hours of this case.
• One other case had a possible epidemiological link to this outbreak through a household
contact, also a laboratory-confirmed case. Sequencing for the household contact is
planned to support these cases being epi-linked to the outbreak.
Cluster membership investigations:
• All samples assigned epidemiologically as part of this cluster are present within this
branch, no further investigations are ongoing.
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 7

 OB-20-108823-AK
Figure 4. Overview of the OB-20-108823-AK
OB-20-108823-AK
Figure 4. Overview of the OB-20-108823-AK
Samples with epidemiological links are highlighted in orange.
Additional cases investigations:
• Genomics indicates links for four additional cases may be part of this outbreak, these
cases are being followed up by PHUs.
Cluster membership investigations:
• Not applicable since only one sequence is available at this point in time.
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 8

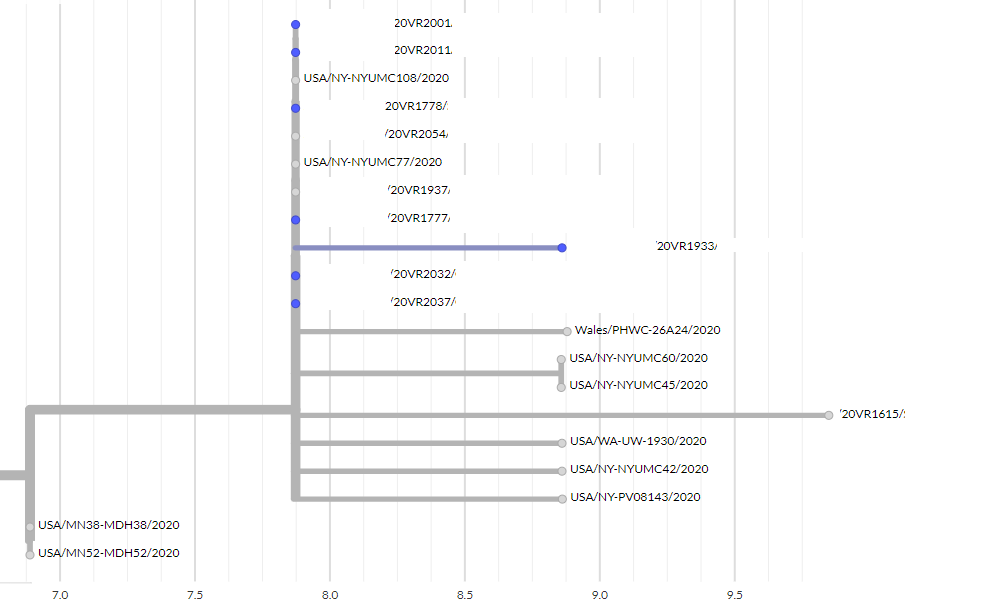
 OB-20-108805-IN (Bluff Wedding)
Figure 5. Overview of the OB-20-108805-IN
OB-20-108805-IN (Bluff Wedding)
Figure 5. Overview of the OB-20-108805-IN
Samples with epidemiological links are highlighted in blue.
Three specimens from one case have been sequenced for quality control purposes.
Additional cases investigations:
• Genomics indicates three additional cases may be linked to this outbreak
• One case has been followed up and may be linked to the outbreak through exposure at a
venue recently visited by some wedding attendees. Further investigations are underway.
• The two other cases have been referred to the public health unit for further investigation.
Cluster membership investigations:
• 20VR**** is epidemiologically linked to the outbreak but three generations from the likely
source case. This may explain why there appears to be one mutation difference from
earlier cases already sequenced. Intermediate cases between the possible source case
and linked to this case have been identified and will be sequenced.
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 9

IDENTIFICATION OF TRANSMISSION PATTERNS
Three different transmission chains identified within one outbreak
Apart from linking cases to known clusters genomic data can also aid investigations into the
relationship of samples within an epidemiological cluster. In the OB-20-108803-AK outbreak
whole genome sequencing identified three unique transmission patterns among cases
epidemiologically assigned to one outbreak. This finding indicates that people that were
considered part of an epidemiological cluster were infected by three unique sources and
require further follow up to review the respective epidemiological links for each transmission
chain. Initial follow up has indicated that there are alternative epidemiological links that could
explain the genomic findings for two of these transmission chains and these cases have
been referred to the public health unit for further investigation.
Table 3. Different transmission chains identified within the OB-20-108803-AK cluster
Route
ESR Lab ID
Outbreak ID
WGS
Link to non-OB case
1
20VR****
OB-20-108803-AK
Yes
1
20VR****
OB-20-108803-AK
Yes
2
20VR****
OB-20-108803-AK
Yes
2
20VR****
-
Yes
20VR****
3
20VR****
OB-20-108803-AK
Yes
OB-20-108820-AK
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 10

GLOSSARY
WGS – Whole Genome Sequencing, the reading of the complete viral RNA genome using
next generation and nanopore sequencing approaches
OB – Outbreak, two or more cases that are epidemiologically linked and not confined to a
single household.
Whole Genome Sequencing of SARS-CoV-2: Progress Report
INSTITUTE OF ENVIRONMENTAL SCIENCE AND RESEARCH LIMITED
Page 11



















